# In individual .qmd files, we would see TheList is modified as seen below
TheList <- c("3L") # Retrieves from 3L archive data folder
TheList <- c("4L") # Retrieves from 4L archive data folder
TheList <- c("5L") # Retrieves from 5L archive data folder
TheList <- c("CS") # Retrieves from CS archive data folderIndividual Instrument Dashboard
Having processed your Instrument QC data for that instrument, when you check the data/instrument/archive folder you will now find both archive data .csv files corresponding to the data for both Gain and MFI tracking. We are now ready to discuss how the dashboard elements are coded and assemble to produce the individual webpages.
For this walk-though, we will be examining the the Aurora5L.qmd file from the original repository.
Walkthrough
When opening the file, the first thing will be the following elements:
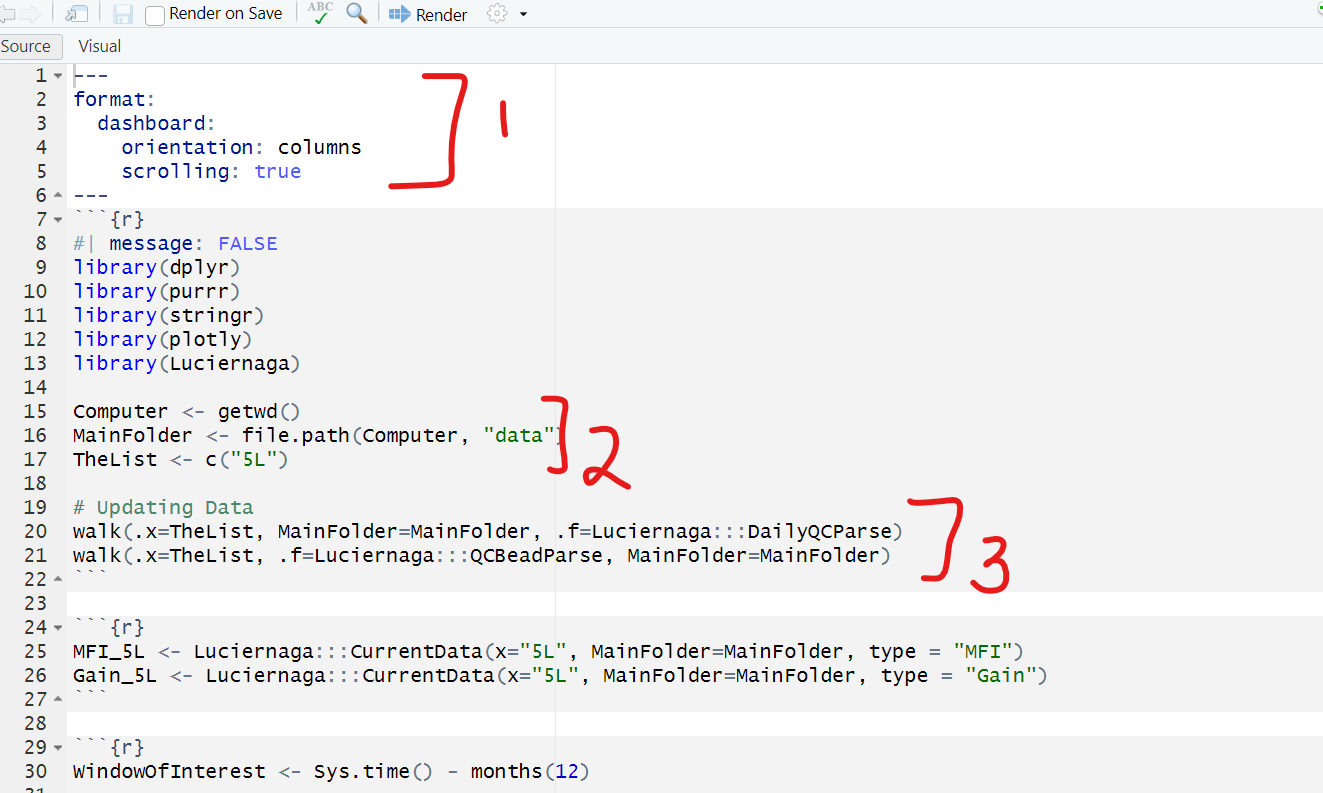 We first see the yaml section. This section specifies to Quarto what kind of document the code is supposed to assemble into. For the instrument pages, it is set up to be a dashboard, with the individual elements set up in sequential columns. The scrolling option is set to TRUE, which allows us to scroll down the page, rather than constraining all plots to the dimensions of the screen.
We first see the yaml section. This section specifies to Quarto what kind of document the code is supposed to assemble into. For the instrument pages, it is set up to be a dashboard, with the individual elements set up in sequential columns. The scrolling option is set to TRUE, which allows us to scroll down the page, rather than constraining all plots to the dimensions of the screen.
We next see a code-block chunk where the necessary packages to execute the code within this .qmd file are loaded via library calls. Then (#2) we have three lines of code that contribute to provide the file.path to the archived processed data folder for the particular instrument. If you were to compare the .qmd files specific for the other instruments (Aurora3L.qmd, Aurora4L.qmd, AuroraCS.qmd) you would find that the code for TheList element is changed within each respective .qmd file, allowing them to retrieve separate stored data.
The next code block is the processing functions that are called in the Rscript, that check to make sure no new QC data has been copied and is waiting to be processed.
Retrieving Data
Having set up the file.path (MainFolder) and specified the particular instrument folder (“5L” in this case) the next two code blocks retrieve the data in the Gain and MFI .csv files, and then filter the data for the last twelve months. If you wanted to modify the time period shown, you would edit the code block at this point to increase/decrease the range.
MFI_5L <- Luciernaga:::CurrentData(x="5L", MainFolder=MainFolder, type = "MFI")
Gain_5L <- Luciernaga:::CurrentData(x="5L", MainFolder=MainFolder, type = "Gain")WindowOfInterest <- Sys.time() - months(12)
MFI_5L <- MFI_5L %>% filter(DateTime >= WindowOfInterest)
Gain_5L <- Gain_5L %>% filter(DateTime >= WindowOfInterest)The following code block references the .csv file containing Field-Service Engineer Visits, which are depicted as red vertical dashed lines in the .pdf version of the plots that can be exported.
Data <- read.csv("AuroraMaintenance.csv", check.names=FALSE)
Data <- Data %>% filter(!str_detect(reason, "lean"))
Repair5L <- Data %>% filter(instrument %in% "5L")Processing the Data
The next three code chunks are rather large, each representing three types of data that will make up the three columns seen on each instruments page (MFI, Gain, RCV). We will work through the first code chunk, which is MFI.
x <- MFI_5L
x <- x %>% dplyr::filter(Timepoint %in% c("Before", "After"))
TheColumns <- x %>% select(where(~is.numeric(.)||is.integer(.))) %>% colnames()
TheColumns <- setdiff(TheColumns, "TIME")
TheIntermediate <- TheColumns[!str_detect(TheColumns, "Gain")]
TheColumnNames <- TheIntermediate[str_detect(TheIntermediate, "-A")]
UltraVioletGains <- TheColumnNames[str_detect(TheColumnNames, "^UV")]
VioletGains <- TheColumnNames[str_detect(TheColumnNames, "^V")]
BlueGains <- TheColumnNames[str_detect(TheColumnNames, "^B")]
YellowGreenGains <- TheColumnNames[str_detect(TheColumnNames, "^YG")]
RedGains <- TheColumnNames[str_detect(TheColumnNames, "^R")]
ScatterGains <- TheIntermediate[str_detect(TheIntermediate, "SC-")]
ScatterGains <- Luciernaga:::ScalePriority(ScatterGains)
LaserGains <- TheIntermediate[str_detect(TheIntermediate, "Laser")]
LaserGains <- Luciernaga:::ColorPriority(LaserGains)
ScalingGains <- TheIntermediate[str_detect(TheIntermediate, "Scaling")]
ScalingGains <- Luciernaga:::ColorPriority(ScalingGains)
OtherGains <- c(ScatterGains, LaserGains, ScalingGains)The retrieved data for MFI goes through a couple processing steps to retrieve the column names present within the .csv file. From there it removes those that show Gain as they will be plotted along with RCV separately. Once this is done, it filters the column names by presence of string characters in their names to separate the list of colnames into shorter list based on laser, scatter, etc.
At this point, each of the above element is simply smaller list of column names, that will then be plotted and visualized together. This happens in the portion of the larger code chunk shown below, with the smaller list being provided to the MeasurementType arguments.
UltraVioletPlotsMFI <- QC_Plots(x=x, FailedFlag=TRUE, MeasurementType=UltraVioletGains,
plotType = "comparison", returntype = "plots",
Metadata="Timepoint", strict = TRUE, YAxisLabel = "MFI",
RepairVisits=Repair5L)
VioletPlotsMFI <- QC_Plots(x=x, FailedFlag=TRUE, MeasurementType=VioletGains,
plotType = "comparison", returntype = "plots",
Metadata="Timepoint", strict = TRUE, YAxisLabel = "MFI",
RepairVisits=Repair5L)
BluePlotsMFI <- QC_Plots(x=x, FailedFlag=TRUE, MeasurementType=BlueGains,
plotType = "comparison", returntype = "plots",
Metadata="Timepoint", strict = TRUE, YAxisLabel = "MFI",
RepairVisits=Repair5L)
YellowGreenPlotsMFI <- QC_Plots(x=x, FailedFlag=TRUE, MeasurementType=YellowGreenGains,
plotType = "comparison", returntype = "plots",
Metadata="Timepoint", strict = TRUE, YAxisLabel = "MFI",
RepairVisits=Repair5L)
RedPlotsMFI <- QC_Plots(x=x, FailedFlag=TRUE, MeasurementType=RedGains,
plotType = "comparison", returntype = "plots",
Metadata="Timepoint", strict = TRUE, YAxisLabel = "MFI",
RepairVisits=Repair5L)
ScatterPlotsMFI <- QC_Plots(x=x, FailedFlag=TRUE, MeasurementType=ScatterGains,
plotType = "comparison", returntype = "plots",
Metadata="Timepoint", strict = TRUE, YAxisLabel = " ",
RepairVisits=Repair5L)
LaserPlotsMFI <- QC_Plots(x=x, FailedFlag=TRUE, MeasurementType=LaserGains,
plotType = "comparison", returntype = "plots",
Metadata="Timepoint", strict = TRUE, YAxisLabel = " ",
RepairVisits=Repair5L)Once this is done, each of the elements above contains a list of plots corresponding to the column names that were provided in the smaller list. These will be referenced later when building the dashboard in the desired layout.
This process is then repeated again in two large code chunks for both Gain and RCV, with some small differences in how the column names are separated into smaller list, and how the individual ggplots are generated.
Starting on line 205 of Aurora5L.qmd we have the following code-chunk:
PDFPlots <- c(UltraVioletPlotsMFI, VioletPlotsMFI, BluePlotsMFI, YellowGreenPlotsMFI, RedPlotsMFI, LaserPlotsMFI, ScatterPlotsMFI, UltraVioletPlotsGain, VioletPlotsGain, BluePlotsGain, YellowGreenPlotsGain, RedPlotsGain, ScatterPlotsGain, LaserDelayPlotsGain, LaserPowerPlotsGain, ScalingPlotsGain, UltraVioletPlotsRCV, VioletPlotsRCV, BluePlotsRCV, YellowGreenPlotsRCV, RedPlotsRCV, ScatterPlotsRCV)
Filename <- paste0("QCPlots_5L")
PDF <- Utility_Patchwork(x=PDFPlots, filename=Filename, returntype="pdf", outfolder=MainFolder, thecolumns=1)This code chunk above assembled all the plots we generated in the section above, and saves them as the .pdf for the individual instrument that can be seen under the data tab of the dashboard website.
Visualizing the Data
MFI
Now that the instrument data has been assembled into plots for the individual measurement types and lasers, it is time to display the plots in a way that produces the instrument page visible on the original dashboard website.
As mentioned at the beginning of this page, the orientation of this dashboard is set to columns. Consequently, on line 218 you will see the following “## MFI {.tabset}”. The two # designate the first column. All plots visualized until the next ## (“## Gain {.tabset}” in our case at line 345) will consequently be present within this first column.
If we desired to change the ordering MFI column last rather and first, we would move everything from “## MFI {.tabset}” until “## Gain {.tabset}” and shift to desired order position.
The presence of “{.tabset}” designates that within this colum (denoted by the ##), there will be multiple tab options to switch between. The first is seen in the following code-chunk:
ggplotly(UltraVioletPlotsMFI[[1]])
ggplotly(UltraVioletPlotsMFI[[2]])
# etc...The tab name visible on the website is denoted by the title: argument. Within the code block itself, the individual plots are made interactive using the plotly function ggplotly, calling the specific detector plot sequentially.
This is then repeated on the next tabs for Violet, Blue, Yellow-Green and Red detectors. Then Scatter, LaserPower, LaserDelay, LaserScatter plots are visualized in their respective tabs.
Finally, marking the end of the first column we find a card element “{.card title=”MFI”}” that appears within the tabset at the end but contains no plots to serve as a reference of what the first column plots are showing.
Gain and rCV
Above we walked through the process of displaying the MFI plots generated in the first section within tabsets for individual lasers. At line 345, we encounter “## Gain {.tabset}” which designates the second column of the dashboard, containing tabs for the Gain plots by laser. The overall layout is similar to what we encountered, differing here and there based on additional plots present within it’s respective archive .csv but not found in the bead csv MFI derrived from.
After all the Gain plots are plotted, we encounter the third column “## RCV {.tabset}” at line 472, continuing until the end.
Summary
The individual instrument .qmd file is setup to retrieve the instrument specific archive data and filter for a desired time range. Once this is done, the individual column names in the .csv are identified and split on the basis of characters in their names into smaller list (typically by laser). This process is then repeated for each measurement type (MFI, Gain, RCV). For the assembly of the actual dashboard itself, the ordering of the measurement type columns is denoted by the ## as encountered, with the tab-sets within each of these also displayed by the order encountered. By rearranging the order, modifications to the individual instrument dashboard pages can be customized.
If you have multiple instruments, you would modify each instruments .qmd file in a similar way to ensure the correct archive data folder is being referenced, and then customize which plots you want displayed when and where. We will discuss this more in the following section.